
PlantDeepSEA
Predicting Regulatory Effect of Genomic Variants
Statistics and Data Evaluation
Statistics of Data Used in PlantDeepSEA
|
Species
|
Reads number
|
Mapping rate | Duplication rate | Mapped reads ( q >30) | TSS enrichment | Peak number | AUROC |
| Bdistachyon_flag_leaf_1 | 94067702 | 0.97 | 0.41 | 49948552 | 13.45 | 61280 | 0.94 |
| Bdistachyon_flag_leaf_2 | 63932496 | 0.96 | 0.34 | 37358896 | 13.10 | 56088 | 0.94 |
| Bdistachyon_flower_1 | 39572726 | 0.95 | 0.58 | 13350236 | 13.73 | 51581 | 0.95 |
| Bdistachyon_flower_2 | 24329952 | 0.96 | 0.55 | 8465850 | 14.75 | 45663 | 0.96 |
| Bdistachyon_panicle_1 | 116923382 | 0.97 | 0.60 | 39842576 | 15.73 | 66431 | 0.95 |
| Bdistachyon_root_1 | 14543028 | 0.47 | 0.15 | 4985401 | 7.47 | 17848 | 0.96 |
| Bdistachyon_root_2 | 45701156 | 0.51 | 0.19 | 16335644 | 6.97 | 46648 | 0.96 |
| Bdistachyon_young_leaf_1 | 10870214 | 0.96 | 0.13 | 8573030 | 9.78 | 32611 | 0.95 |
| Bdistachyon_young_leaf_2 | 10291646 | 0.98 | 0.12 | 7935817 | 8.15 | 25245 | 0.95 |
| MH63_flag_leaf_1 | 126342026 | 0.99 | 0.29 | 67822335 | 9.10 | 83771 | 0.93 |
| MH63_flag_leaf_2 | 64745662 | 0.99 | 0.23 | 30410515 | 6.83 | 58225 | 0.95 |
| MH63_flower_2c_1 | 85892532 | 0.99 | 0.16 | 57881493 | 7.70 | 91919 | 0.97 |
| MH63_flower_2c_2 | 122252060 | 0.99 | 0.25 | 75131980 | 9.22 | 97974 | 0.98 |
| MH63_lemma_1 | 74239198 | 0.99 | 0.24 | 48165596 | 10.19 | 92319 | 0.95 |
| MH63_lemma_2 | 85247604 | 0.99 | 0.21 | 56179824 | 9.81 | 96716 | 0.95 |
| MH63_panicle_1 | 58832756 | 0.98 | 0.44 | 24165690 | 14.84 | 66847 | 0.96 |
| MH63_panicle_2 | 33309586 | 0.99 | 0.35 | 14620120 | 15.59 | 54404 | 0.97 |
| MH63_panicle_3 | 51162146 | 0.99 | 0.36 | 21968846 | 15.72 | 62186 | 0.97 |
| MH63_panicle_4 | 55158396 | 0.98 | 0.34 | 25736079 | 11.75 | 71983 | 0.96 |
| MH63_panicle_5 | 46253072 | 0.99 | 0.31 | 25128785 | 11.95 | 72647 | 0.96 |
| MH63_root_1 | 223711752 | 0.98 | 0.63 | 66247510 | 15.61 | 83155 | 0.95 |
| MH63_root_2 | 68284992 | 0.99 | 0.44 | 31913324 | 14.21 | 52070 | 0.96 |
| MH63_young_leaf_1 | 138938116 | 0.99 | 0.51 | 44111400 | 13.01 | 64920 | 0.95 |
| MH63_young_leaf_2 | 70109302 | 0.99 | 0.43 | 27587710 | 14.21 | 52070 | 0.95 |
|
ZS97_flag_leaf_1
|
92533216 | 0.98 | 0.28 | 49541050 | 14.77 | 79501 | 0.95 |
| ZS97_flag_leaf_2 | 163670488 | 0.97 | 0.62 | 52962753 | 14.22 | 77310 | 0.95 |
| ZS97_flower_1 | 101666618 | 0.98 | 0.25 | 65770237 | 12.84 | 109599 | 0.94 |
| ZS97_flower_2 | 98606866 | 0.99 | 0.24 | 63957818 | 13.18 | 99573 | 0.94 |
| ZS97_lemma_1 | 37055650 | 0.99 | 0.15 | 25899484 | 11.66 | 84317 | 0.96 |
| ZS97_lemma_2 | 38347774 | 0.99 | 0.17 | 26442996 | 12.02 | 81764 | 0.96 |
| ZS97_panicle_1 | 57620486 | 0.97 | 0.37 | 28078266 | 14.38 | 65456 | 0.96 |
| ZS97_panicle_2 | 24321634 | 0.99 | 0.32 | 10985233 | 13.95 | 51842 | 0.97 |
| ZS97_panicle_3 | 36435606 | 0.98 | 0.35 | 15184026 | 13.70 | 55265 | 0.96 |
| ZS97_panicle_4 | 57332524 | 0.99 | 0.28 | 28415206 | 10.31 | 66366 | 0.96 |
| ZS97_panicle_5 | 65002916 | 0.99 | 0.36 | 28751790 | 10.72 | 66250 | 0.96 |
| ZS97_root_1 | 166991452 | 0.98 | 0.58 | 52947523 | 15.53 | 83093 | 0.95 |
| ZS97_root_2 | 74643946 | 0.99 | 0.43 | 33806877 | 15.76 | 73842 | 0.95 |
| ZS97_young_leaf_1 | 174433758 | 0.98 | 0.60 | 45111050 | 17.78 | 62008 | 0.95 |
| ZS97_young_leaf_2 | 28235594 | 0.98 | 0.38 | 11624830 | 17.22 | 35404 | 0.96 |
| Sitalica_flag_leaf_1 | 141996604 | 0.99 | 0.25 | 85826001 | 6.06 | 65251 | 0.95 |
| Sitalica_flag_leaf_2 | 100706220 | 0.99 | 0.21 | 63590531 | 6.29 | 58482 | 0.95 |
| Sitalica_flower_1 | 131086536 | 0.99 | 0.23 | 91347631 | 7.20 | 95685 | 0.95 |
| Sitalica_flower_2 | 138357024 | 0.99 | 0.21 | 98776879 | 7.34 | 89544 | 0.95 |
| Sitalica_panicle_1 | 177674200 | 0.99 | 0.20 | 125600542 | 6.39 | 101296 | 0.95 |
| Sitalica_panicle_2 | 150142666 | 0.99 | 0.23 | 100278310 | 6.38 | 93938 | 0.95 |
| Sitalica_root_2c_1 | 25735750 | 0.90 | 0.56 | 7811308 | 7.48 | 32704 | 0.98 |
| Sitalica_young_leaf_2c_1 | 70067138 | 0.97 | 0.44 | 29928285 | 8.26 | 70194 | 0.97 |
| Sitalica_young_leaf_2c_2 | 57397792 | 0.98 | 0.56 | 19724755 | 9.74 | 41994 | 0.96 |
| Sorghum_flag_leaf_1 | 130334958 | 0.99 | 0.44 | 52409705 | 11.24 | 56465 | 0.98 |
| Sorghum_flag_leaf_2 | 144450460 | 0.99 | 0.41 | 60226969 | 10.82 | 70472 | 0.98 |
| Sorghum_flower_1 | 203189918 | 1.00 | 0.16 | 133020280 | 7.67 | 142580 | 0.97 |
| Sorghum_flower_2 | 245343676 | 1.00 | 0.15 | 159542821 | 7.32 | 159156 | 0.97 |
| Sorghum_lemma_1 | 95665980 | 1.00 | 0.17 | 65182860 | 8.99 | 104655 | 0.97 |
| Sorghum_lemma_2 | 169146264 | 1.00 | 0.16 | 114027776 | 7.67 | 115553 | 0.97 |
| Sorghum_panicle_bottom_1 | 84595194 | 0.98 | 0.32 | 38245112 | 7.69 | 80219 | 0.97 |
| Sorghum_panicle_bottom_2 | 182882314 | 0.99 | 0.56 | 52812432 | 8.01 | 87226 | 0.97 |
| Sorghum_panicle_top_1 | 37071094 | 0.85 | 0.28 | 16029710 | 8.17 | 49753 | 0.98 |
| Sorghum_panicle_top_2 | 39628252 | 0.99 | 0.31 | 19196228 | 8.86 | 57743 | 0.98 |
| Sorghum_root_2c_1 | 63639878 | 0.87 | 0.52 | 10997618 | 10.90 | 50778 | 0.99 |
| Sorghum_root_2c_2 | 397222962 | 0.99 | 0.78 | 60817144 | 12.39 | 83349 | 0.97 |
| Sorghum_young_leaf_2c_1 | 112887230 | 0.96 | 0.42 | 29067302 | 15.35 | 60549 | 0.98 |
| Sorghum_young_leaf_2c_2 | 8595822 | 0.99 | 0.15 | 5260625 | 13.03 | 32671 | 0.99 |
| Zmays_ear_big_bottom_1 | 92636560 | 1.00 | 0.32 | 40646406 | 10.99 | 79423 | 0.98 |
| Zmays_ear_big_bottom_2 | 96513184 | 0.99 | 0.36 | 38565782 | 11.25 | 80683 | 0.98 |
| Zmays_ear_big_top_1 | 96724598 | 1.00 | 0.34 | 40941794 | 10.56 | 78920 | 0.98 |
| Zmays_ear_big_top_2 | 101609846 | 0.99 | 0.37 | 38880627 | 10.25 | 75897 | 0.98 |
| Zmays_ear_small_1 | 91987754 | 0.99 | 0.39 | 36185687 | 11.05 | 78159 | 0.98 |
| Zmays_ear_small_2 | 96011440 | 0.99 | 0.31 | 42669858 | 11.05 | 88322 | 0.98 |
| Zmays_flag_leaf_1 | 95249354 | 1.00 | 0.37 | 43666323 | 8.55 | 52096 | 0.98 |
| Zmays_flag_leaf_2 | 100750444 | 1.00 | 0.43 | 39967001 | 9.49 | 63572 | 0.98 |
| Zmays_flower_1 | 105904206 | 0.99 | 0.48 | 21233571 | 5.37 | 24667 | 0.98 |
| Zmays_flower_2 | 112607990 | 1.00 | 0.29 | 55306202 | 5.53 | 26295 | 0.99 |
| Zmays_root_2c_1 | 226773650 | 0.50 | 0.29 | 40789638 | 15.08 | 98405 | 0.98 |
| Zmays_root_2c_2 | 158419674 | 0.43 | 0.29 | 22939197 | 14.79 | 82732 | 0.99 |
| Zmays_root_2c_3 | 159349756 | 1.00 | 0.32 | 80542696 | 11.95 | 90462 | 0.98 |
| Zmays_tassel_bottom_1 | 94275220 | 1.00 | 0.30 | 42490103 | 8.87 | 56771 | 0.98 |
| Zmays_tassel_bottom_2 | 86178832 | 1.00 | 0.35 | 35440479 | 9.71 | 62743 | 0.98 |
| Zmays_tassel_top_1 | 180483274 | 1.00 | 0.42 | 71876863 | 9.08 | 70640 | 0.98 |
| Zmays_young_leaf_2c_1 | 141333474 | 0.95 | 0.34 | 41835977 | 16.62 | 90765 | 0.99 |
| Zmays_young_leaf_2c_2 | 103976508 | 0.95 | 0.29 | 30775805 | 15.40 | 75124 | 0.99 |
| Zmays_young_leaf_2c_3 | 126330162 | 0.97 | 0.40 | 57470896 | 13.05 | 82855 | 0.98 |
|
Arabidopsis_7days_leaf_rep1
|
45,225,502 | 99.49% | 30.40% | 13,461,588 | 12.32 | 25,571 | 0.92 |
| Arabidopsis_7days_leaf_rep2 | 149,379,644 | 71.71% | 30.00% | 30,345,758 | 15.54 | 33,398 | 0.94 |
| Arabidopsis_root_tip_rep1 | 47,952,800 | 98.71% | 6.04% | 29,813,691 | 5.31 | 17,272 | 0.95 |
| Arabidopsis_root_tip_rep2 | 121,576,490 | 98.58% | 8.50% | 73,908,585 | 3.98 | 10,784 | 0.95 |
| Arabidopsis_root_non_hair_cell_rep1 | 94,079,852 | 99.49% | 10.17% | 60,213,985 | 11.19 | 34,494 | 0.95 |
| Arabidopsis_root_non_hair_cell_rep2 | 24,296,952 | 99.36% | 5.79% | 15,406,787 | 9.94 | 23,224 | 0.96 |
| Arabidopsis_root_hair_cell_rep1 | 109,162,074 | 99.30% | 10.40% | 68,423,950 | 9.62 | 33,853 | 0.94 |
| Arabidopsis_root_hair_cell_rep2 | 29,171,548 | 99.38% | 6.42% | 18,573,026 | 10.88 | 26,433 | 0.95 |
| Arabidopsis_stem_cell_rep1 | 84,687,814 | 99.49% | 21.81% | 27,662,167 | 14.57 | 29,395 | 0.94 |
| Arabidopsis_stem_cell_rep2 | 99,733,622 | 99.51% | 20.35% | 36,838,477 | 14.40 | 31,004 | 0.96 |
| Arabidopsis_stem_cell_rep3 | 151,660,948 | 99.53% | 25.65% | 42,330,835 | 13.97 | 30,946 | 0.94 |
| Arabidopsis_mesophyll_cell_rep1 | 112,729,514 | 99.14% | 32.76% | 12,581,426 | 15.35 | 21,455 | 0.95 |
| Arabidopsis_mesophyll_cell_rep2 | 99,938,318 | 99.32% | 28.60% | 18,268,744 | 16.10 | 23,916 | 0.94 |
| Arabidopsis_mesophyll_cell_rep3 | 100,129,142 | 99.27% | 25.78% | 10,905,730 | 16.10 | 21,512 | 0.96 |
| Arabidopsis_mesophyll_cell_rep3 | 100,129,142 | 99.27% | 25.78% | 10,905,730 | 16.10 | 21,512 | 0.96 |
Quality Control
In order to obtain more accurate training results, we use a series of quality control measures to ensure the accuracy of the deep learning model input data.
Principal component analysis (PCA)
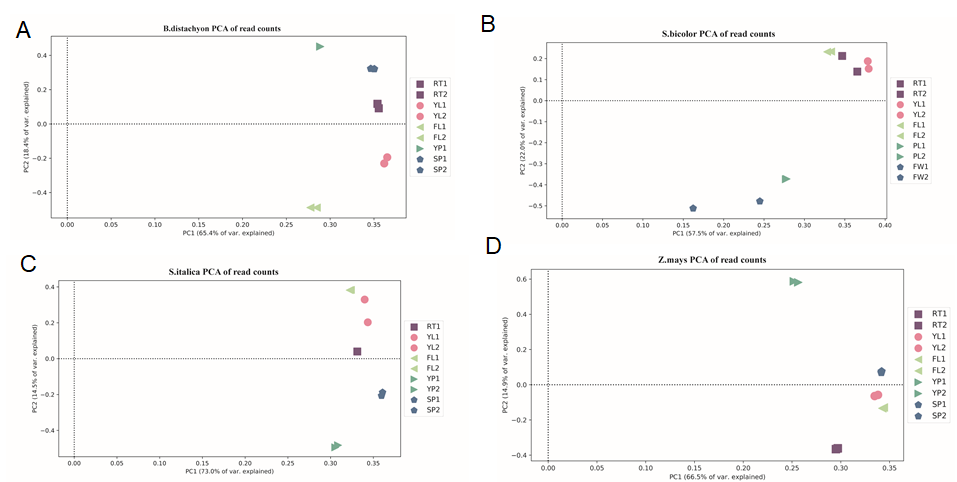
Figure 1. PCA plot of of read counts for each library shows that biological replicates of samples in Brachypodium distachyon (A), Sorghum bicolor (B), Setaria italica (C) and Zea mays (D). RT, root; YL, young leaf; FL, flag leaf; YP, young panicle; SP, stamen & pistil.
Enrichment of ATAC-seq reads centered at TSSs (transcription start sites)
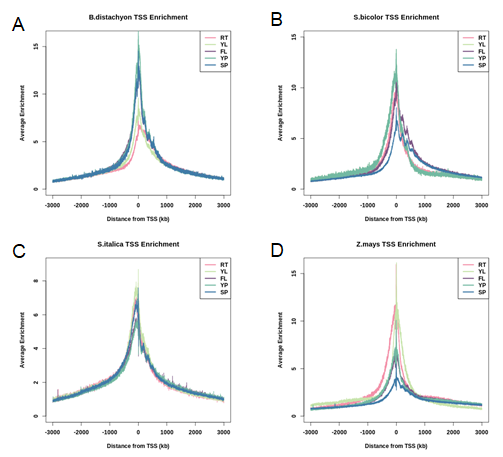
Figure 2. Enrichment of ATAC-seq reads centered at TSSs, cut sites number is normalized by quantile normalization at regions flanking the TSS (from −3000 to +3000) of samples in Brachypodium distachyon (A), Sorghum bicolor (B), Setaria italica (C) and Zea mays (D). RT, root; YL, young leaf; FL, flag leaf; YP, young panicle; SP, stamen & pistil.
Evaluation of Deep Learning Models
Receiver operating characteristic (ROC)
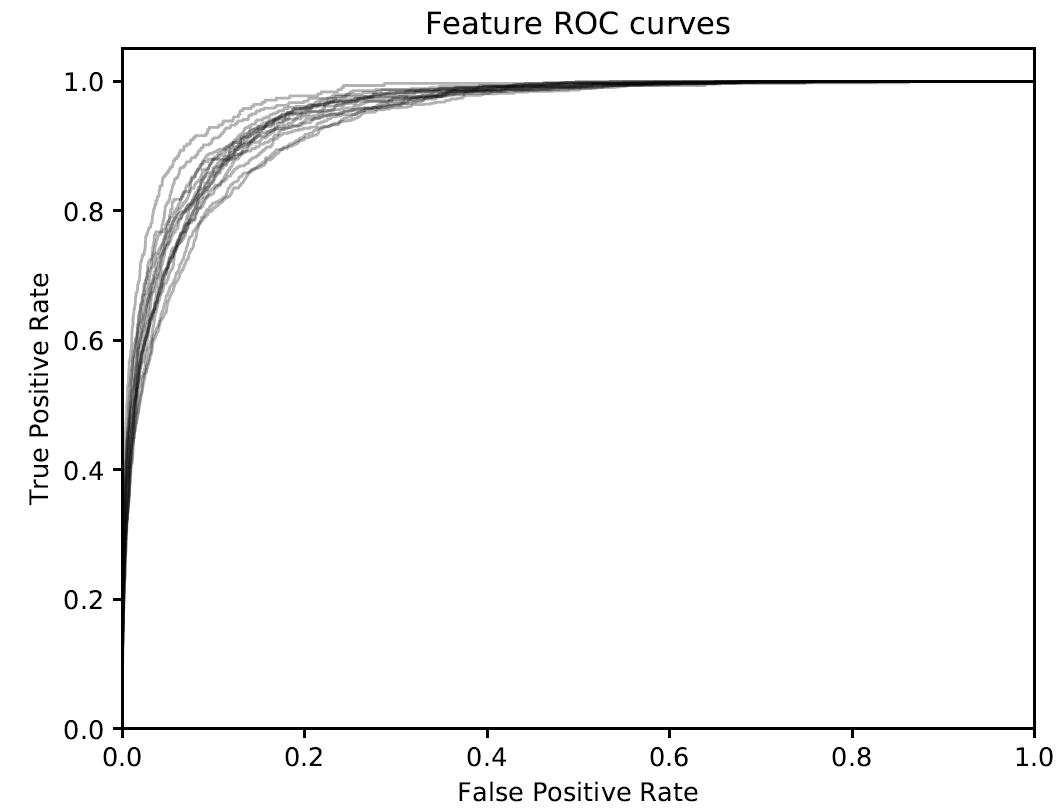
Figure 3. Receiver operating characteristic (ROC) curve for different samples of rice cultivar Zhenshan97.